How to predict the basic chemical properties?
The TargetNet can only be accessed by selecting 'Basic Properties Prediction' link shown as the following figure.

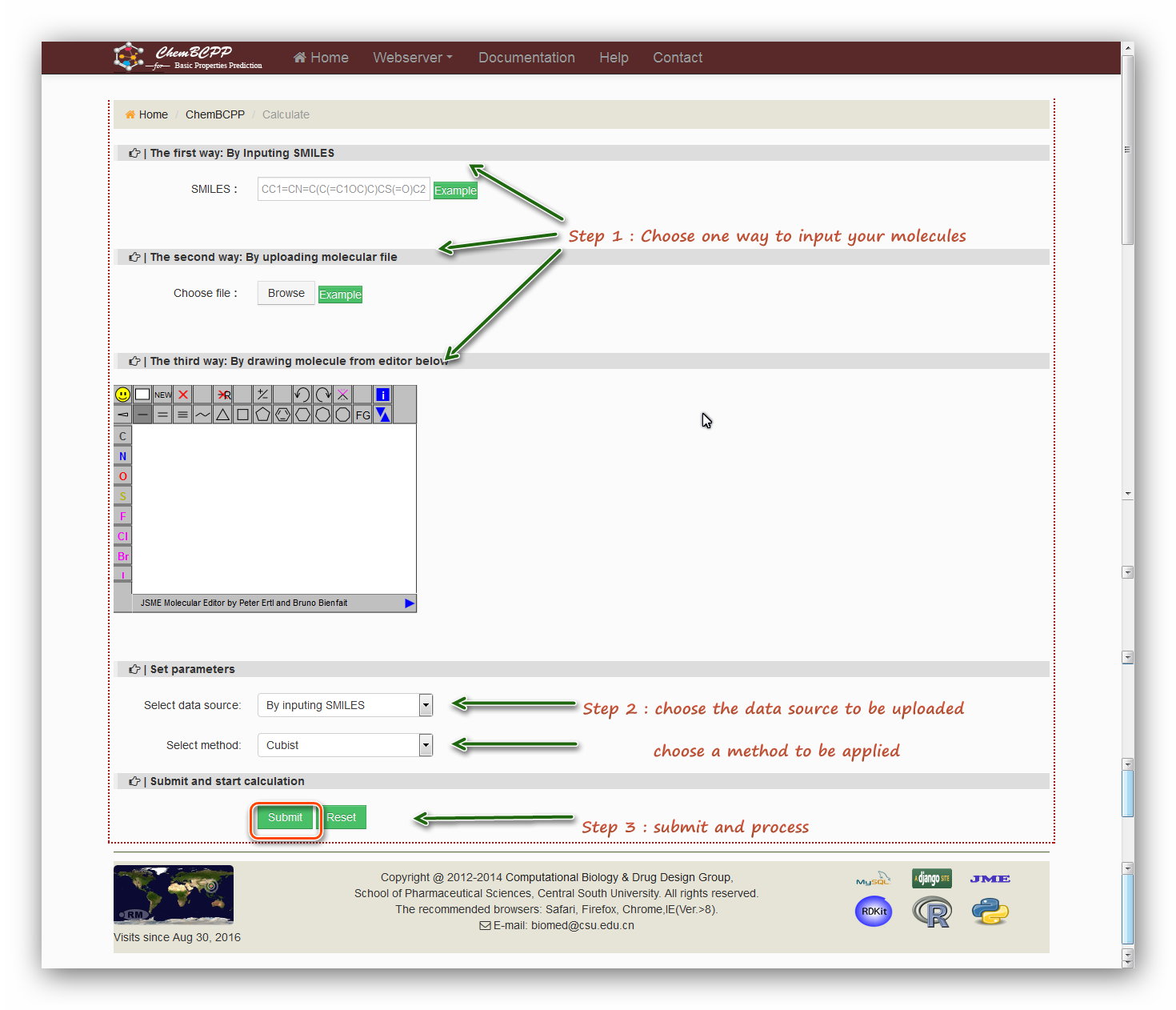
Step 1: Users are required to submit a molecular structure in SMILES format or a molecular file in SDF format or to draw a structure using the JME editor.
The first way: By Inputing SMILES
Type in SMILES into the field or click the example button to load a example SMILES.
The second way: By uploading molecular file
Choose a molecular file in format of SDF.
Users can also click the ‘example’ button to download a molecular file for an example.
The third way: By drawing molecule from editor below
Input a molecule by drawing structure in the JSME editor form Peter Ertl.
Step 2: Set parameters
There are two parameters need to be set. The first one: select data source can be chosen according to the step 1. The second parameter: select method can
be chosen from 'Ramdom forest', 'Cubist', 'SVM', and 'Boosting' which are four algorithms applied into the models for prediction.
Step 3: Submit and process
Click the 'Submit' button and then start processing. The processing will last for a while and then users will be redirected to the result page.
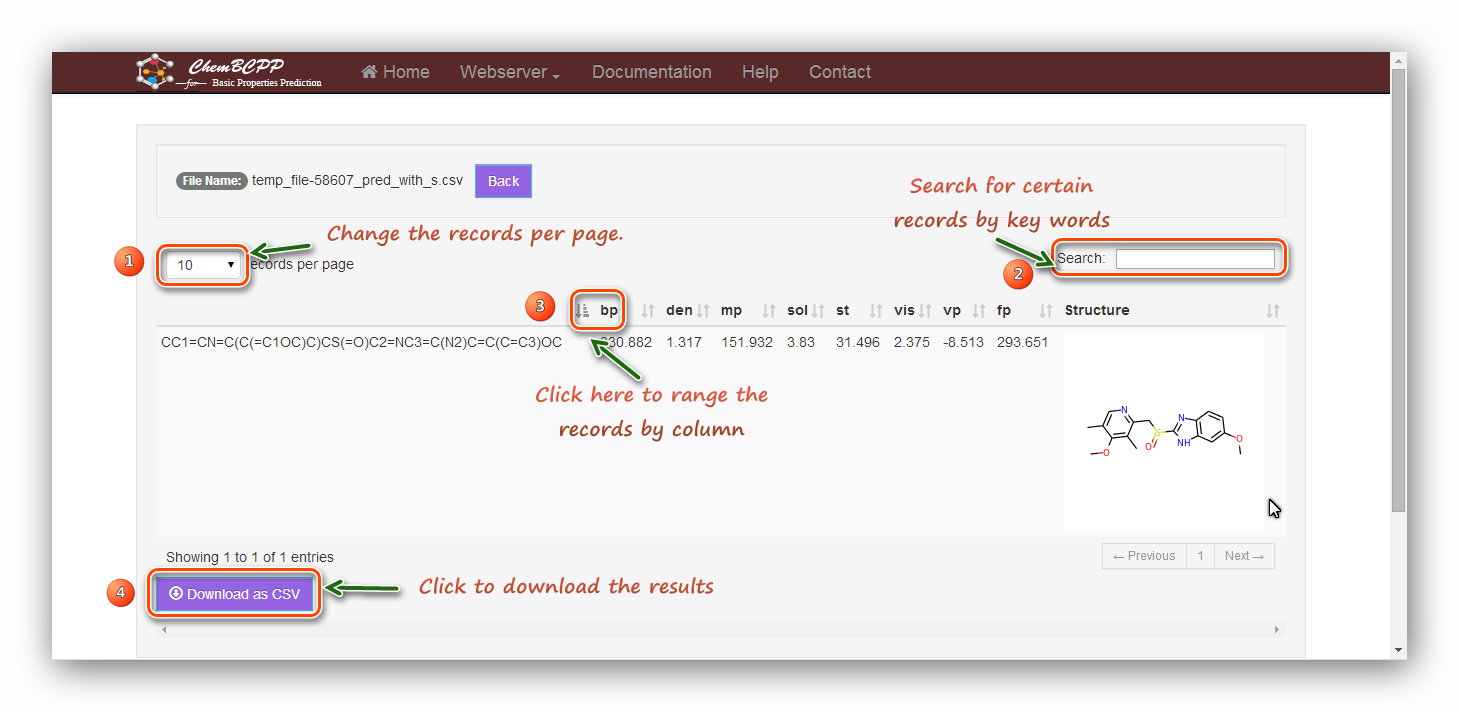
Step 4: Results
The result table contains
"bp", "den", "mp", "sol", "st", "vis", "vp", and "fp". "1" marks the ‘per page’ button, and users can choose the number of items shown in the table. "2" marks the "search" button, and users can conveniently type in a keyword to
look for a certain item in the results. "3" marks the ‘rank’ button, and users can click the button to get an increasing or decreasing order
of the values. "4" marks the "download" button, and users can download the results as a *.csv file.